The most common method for oligonucleotide synthesis is using the cyanoethyl phosphoramidite to form the backbone structure of DNA and continuing to connect the phosphodiester bonds through “phosphite triester method” (Nucl. Acids Res. 1984, 12, 4539; Tetrahedron Lett. 1983, 24,5843). This makes it possible to synthesize oligonucleotides with high efficiency in a short time (synthesis efficiency > 98%). Long term storage is possible as phosphoramidite monomers are already stabilized before being activated for coupling process.
The synthesis process begins with a solid support attached with a nucleoside. Cycles of deblocking, coupling, oxidation, and capping is repeated to attain the desired sequence of oligonucleotides. Refer to the following figure.
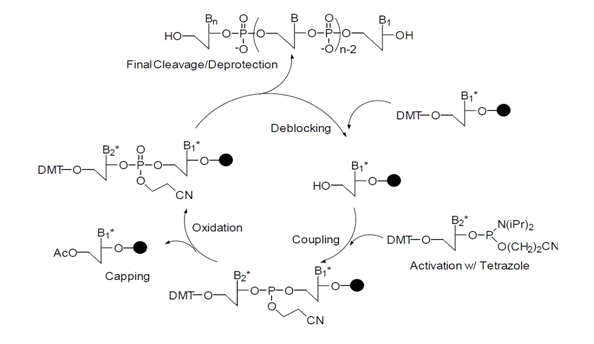
(1) Deblocking
The deblocking process, the first step in the synthesis cycle, involves a reaction to remove DMT group, a protecting group for 5’OH base attached to a solid support. For this to happen, an acidic condition is necessary, and 3% trichloroacetic acid is mostly used. However, some reports claim that acidity may cause depurination, which the bonds between a purine base and a sugar ring are broken, especially when the base is adenosine. Furthermore, trichloroacetic acid is a very strong acid (pKa: ~ 1.5). If it is used for the deblocking process, one must be cautious not to leave the reaction for a long time, or else the depurination will occur. To avoid this problem, in some cases, dichloroacetic acid, a weak acid, may be used instead. During deblocking, the DMT positive ion leaving the solid support are dark orange in color, and their absorbance can be used to measure binding efficiency during oligo synthesis.
(2) Coupling
The coupling reaction is done with the nucleoside phosphoramidite monomers and 5’-hydroxyl groups produced during the deblocking process on the solid support to synthesize the oligonucleotide having the desired sequence. The amine group of nucleoside phosphoramidite monomers used in this step is protected with benzoyl groups (for adenosine and cytidine) or isobutyryl groups (for guanosine), while 5’-hydroxyl groups of all the monomers are protected with DMT. Those serve as bridges during the coupling process. Refer to the following figure.
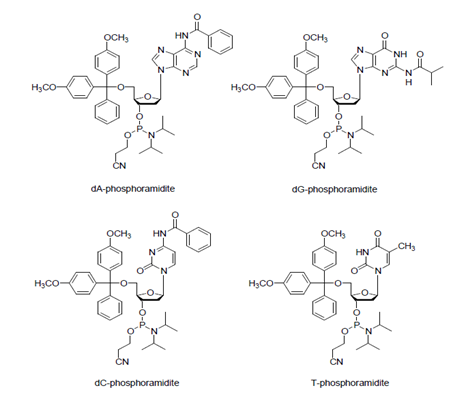
As the phosphoramidite used in this process is already in stabilized structure, those must first undergo activation process to bind with the 5’ hydroxyl group on the solid support. Tetrazole is normally used as an activator to react with phosphoramidite to protonate the nitrogen and convert diisopropyl amino groups into a highly reactive tetrazolide structure.
This results in the formation of phosphite triester bonds from the combination reaction between the tetrazolide and the 5’-hydroxyl group of the solid support. The high reactive nature of the tetrazolide structure will form unwanted structures in the presence of water, even if it exists in a small amount. Thus, having anhydrous conditions are highly important in the coupling process.
(3) Oxidation
The phosphite triester structure formed by the coupling of phosphoramidite and 5’-hydroxyl group must be stabilized by converting it to phosphate triester structure. This is done through an oxidation reaction using an iodine.
(4) Capping
Since the coupling reaction cannot occur quantitatively (usually > 98%), some 5’-hydroxyl groups remain unreacted on the solid support. If those oligos are carried to the coupling step of the next cycle, those will result in having the sequence of one less amino acid, with those being (N-1) mer long.
Therefore, it is important to remove them after the synthesis process, as they make the purification process to get the desired oligonucleotides difficult, making the ‘capping’ process necessary on the 5’ hydroxyl groups to prevent their further reaction. For this, acetylation must be done using acetic anhydride and N-methylimidazole.
Oligonucleotide synthesis of desired length is accomplished by repeating the above procedures. Afterwards, oligonucleotides are treated with ammonia to purify and isolate the oligonucleotides.