Performing gene to protein synthesis in a cell-free system offers a unique approach compared to relying on living cells. Here's a breakdown of the key steps involved:
- 1. Preparation of Cell-Free Extract:
- The first step involves creating a cell-free extract. This is essentially a cellular lysate, a suspension made by breaking open cells and removing membranes and cellular debris.
- The extract retains the essential machinery necessary for protein synthesis, including ribosomes, transfer RNAs (tRNAs), amino acids, and enzymes required for translation initiation, elongation, and termination.
- Common sources for cell-free extracts include E. coli, wheat germ, and rabbit reticulocyte lysates, each with advantages and limitations depending on the desired protein.
- 2. DNA Template Acquisition:
- You'll need a DNA template encoding the protein of interest. Here are two main approaches:
- Plasmids: These circular DNA molecules can be engineered to contain the gene sequence along with regulatory elements for efficient transcription in the cell-free system. Plasmids are often used when large quantities of protein are needed.
- PCR Products: PCR can be used to amplify a specific DNA fragment containing the desired gene sequence. This approach is faster than plasmid construction but might yield lower protein amounts.
- 3. In Vitro Transcription (IVT):
- This step involves synthesizing mRNA from the DNA template. Purified RNA polymerase enzymes are introduced into the cell-free extract along with the DNA template and nucleoside triphosphates (the building blocks of RNA).
- The RNA polymerase recognizes the promoter sequence on the DNA and uses it to transcribe the gene into a complementary mRNA molecule. This mRNA molecule carries the genetic information for protein synthesis.
- 4. Cell-Free Translation:
- The synthesized mRNA is added to the cell-free extract containing all the necessary components for translation.
- Ribosomes in the extract recognize the start codon (AUG) on the mRNA and initiate protein synthesis. tRNAs carrying specific amino acids interact with the mRNA codons, and the ribosome links the amino acids together according to the mRNA sequence.
- This process continues until a stop codon is reached on the mRNA, signaling the release of the completed protein from the ribosome.
Advantages of Cell-Free Protein Synthesis:
- Speed and Simplicity: Cell-free systems offer a faster and more streamlined approach compared to traditional methods involving cell culture.
- Scalability: Protein production can be easily scaled up or down by adjusting the reaction volume and components.
- Purity: Proteins synthesized in cell-free systems are typically free of contaminants present in whole cells.
- Direct Manipulation: The system allows for easy manipulation of reaction conditions and incorporation of non-standard amino acids into the protein.
Applications of Cell-Free Protein Synthesis:
- Rapid Protein Production: This technique is valuable for research purposes, such as studying protein function or screening for protein activity.
- Membrane Protein Production: Cell-free systems can be advantageous for producing membrane proteins, which are often difficult to express in living cells.
- In Vitro Diagnostics: Cell-free protein synthesis can be used to develop diagnostic tools by expressing specific antigens for antibody detection.
- Biomanufacturing: This approach holds promise for the future production of complex proteins for various applications.
Q1: What is Codon Optimization, and why is it necessary?
Codon optimization is the process of strategically modifying the DNA sequence of a gene—without changing the amino acid sequence it encodes—to improve its expression efficiency, stability, synthesizability, and overall performance. It's a crucial synthetic biology technique that goes beyond simple codon changes. Bioneer's GeneCrafter™ is a powerful tool designed for this purpose.
Why is optimization necessary?
- Overcoming Codon Usage Bias (CUB):
- Most amino acids can be coded by multiple codons (DNA triplets), but organisms often show a preference for using certain codons over others. This pattern is known as Codon Usage Bias (CUB). For example, while UCU, UCC, UCA, and UCG all code for Serine, an organism might primarily use only one or two of these efficiently.
- When expressing a gene from one organism (like a human) in a different host system (like E. coli), differences in CUB can drastically slow down or stall mRNA translation, leading to low protein yields. GeneCrafter™ optimizes codons to match the CUB of the target host, maximizing translation efficiency.
- Eliminating Problematic Sequences & Enhancing Functionality:
- Repeat Sequences: Simple repeats (Homopolymers, e.g., AAAAAA) or more complex repeats can cause errors during gene synthesis or lead to genetic instability (e.g., recombination) within the host.
- mRNA Secondary Structures: mRNA can fold back on itself, forming structures like hairpins. These structures, especially near the 5' end, can block ribosome access, hindering translation initiation or slowing down elongation.
- Restriction Enzyme Sites: The presence of unwanted restriction sites can complicate downstream cloning steps or the assembly of genetic circuits.
- CpG/UpA Dinucleotides: Particularly relevant for mRNA vaccines and therapeutics, these dinucleotides can trigger innate immune responses or promote mRNA degradation.
- Other Regulatory Motifs: Undesired sequences like cryptic splice sites, internal prokaryotic ribosome binding sites (RBS), premature polyadenylation signals, or transcription factor binding sites (TFBS) within the coding region can lead to unintended processing or regulation.
- Gene sequences can contain various hidden elements that negatively impact synthesis or expression:
- GeneCrafter™ identifies and removes these problematic sequences, improving gene synthesis success rates, ensuring predictable gene expression, and enabling stable function. It can also be used for attenuation (weakening expression or function) by strategically modifying sequences, which is useful in applications like vaccine development.
Benefits of GeneCrafter™ Optimization:
- Predictable and High Levels of Gene Expression: Maximizes protein or mRNA yield through host-optimized codons and stable mRNA structures.
- Improved mRNA Stability and Functionality: Enhances In Vitro Transcription (IVT) efficiency, increases translation efficiency, and boosts the efficacy of mRNA-based drugs (vaccines, therapeutics) by removing destabilizing elements and optimizing structure.
- Higher Gene Synthesis Success Rate & Cost-Effectiveness: Facilitates faster and more reliable gene synthesis by eliminating sequence complexities (GC imbalances, repeats, hairpins), often leading to more competitive pricing.
- Essential for Heterologous Expression: Overcomes expression barriers between different species, ensuring genes function reliably in the desired host system.
Q2: Can I get gene optimization services using GeneCrafter™?
Bioneer offers custom Codon Optimization and Attenuation services free of charge, powered by our proprietary GeneCrafter™ software.
To request this service, please email the following information to geneorder@bioneer.co.kr:
- DNA or Protein Sequence: The sequence you want to optimize (FASTA format preferred).
- Target Host/Organism: The organism where the gene will be expressed (e.g., E. coli K12, Homo sapiens, CHO-K1, S. cerevisiae).
- Restriction Enzymes to Avoid: List any enzyme sites that should be removed for your downstream cloning or experiments (Optional).
- Primary Application: The main goal of optimization (e.g., Protein expression, mRNA production, Vaccine development).
- CAI Attenuation: Specify if you need to intentionally reduce translation efficiency (Optional, specify desired level if applicable).
Q3: What is GeneCrafter™?
GeneCrafter™ is Bioneer's in-house developed, state-of-the-art software for codon optimization and attenuation. It employs sophisticated Multi-Objective Optimization algorithms that go far beyond simple codon replacement. GeneCrafter™ considers a wide range of biological and physicochemical factors influencing gene expression, stability, and synthesis efficiency to deliver superior, tailor-made gene designs.
Q4: What's the difference between traditional codon optimization and GeneCrafter™?
Traditional codon optimization often focuses on a single objective: matching the Codon Usage Bias (CUB) of the target host to improve translation efficiency.
GeneCrafter™, however, utilizes a far more comprehensive and sophisticated Multi-Objective Optimization approach. Think of it like conducting an orchestra versus playing a single instrument. GeneCrafter™ simultaneously considers and balances multiple critical parameters to achieve the best overall gene performance:
- Codon Usage Bias (CUB): (Fundamental) Uses codons preferred by the host to enhance translation efficiency. GeneCrafter™ can also factor in codon context effects, not just individual frequencies.
- GC Content: (Stability & Synthesis) Modulates both overall and local GC content within optimal ranges to prevent issues caused by excessively stable (high GC) or unstable (low GC) DNA/mRNA regions, improving synthesis success rates.
- mRNA Secondary Structure: (Efficiency & Stability) Minimizes the formation of detrimental structures like hairpins that can impede translation, particularly optimizing the translation initiation site region for better ribosome accessibility.
- Sequence Motifs to Avoid: (Functionality & Safety) Eliminates sequences that could interfere with gene function or downstream processes, ensuring predictability and flexibility:
- Restriction Enzyme Sites: Preserves cloning options.
- Instability Elements: (e.g., AU-rich elements - AREs) Prevents premature mRNA degradation.
- Aberrant Processing Signals: (e.g., Cryptic Splice Sites, Prokaryotic RBS in eukaryotes) Avoids unintended gene regulation or processing.
- Repeat Sequences: (Stability & Synthesis) Removes homopolymers, tandem repeats, and complex internal repeats to reduce gene synthesis errors and potential genetic instability.
- Codon Pair Bias (CPB): (Translation Speed & Accuracy) Considers the influence of adjacent codon pairs on translation speed and fidelity, allowing for fine-tuning of the translation process and potentially impacting protein folding.
- Reduced Immunogenicity: (Safety - especially for mRNA) Crucial for in vivo applications like mRNA vaccines/therapeutics. Minimizes unwanted innate immune responses by modulating CpG and UpA dinucleotide frequencies, removing dsRNA-forming inverted repeats, and avoiding known immune-stimulatory motifs (e.g., certain TLR agonist sequences).
In essence, GeneCrafter™ moves beyond basic CUB matching to provide a robust, reliable solution for synthetic biology challenges, proactively addressing potential issues across the entire Design-Build-Test cycle.
Bioneer's ExiProgen™ is a system specifically designed for automated protein synthesis and nucleic acid extraction. It simplifies and streamlines these processes within a research environment.
- ExiProgen utilizes cell-free protein synthesis. This means it does not rely on living cells but uses extracts containing the essential elements for protein production.
- The system is fully automated:
- Add required reagents and kit components.
- Inject a DNA template (plasmid or PCR product) encoding the protein of interest.
- ExiProgen then handles the entire process of generating mRNA from the DNA template through IVT and then translating the mRNA into protein.
A cell-free system is a powerful tool used in biotechnology and research that essentially mimics protein synthesis outside of a living cell. Here's a breakdown of the concept:
Traditional Protein Synthesis:
- In living cells, protein synthesis occurs within the cytoplasm using the complex machinery found inside the cell.
- This process involves two key steps:
- Transcription (DNA to mRNA): DNA in the nucleus serves as the blueprint. An enzyme called RNA polymerase copies the DNA sequence into a single-stranded messenger RNA (mRNA) molecule.
- Translation (mRNA to Protein): mRNA carries the genetic instructions to ribosomes, which are protein-building factories in the cytoplasm. Ribosomes use the mRNA code to assemble amino acids into a specific protein sequence.
Cell-Free Systems:
- A cell-free system bypasses the need for whole cells. It provides a simplified environment containing the essential cellular components required for protein synthesis in vitro (literally, "in glass").
- These components are typically derived from a cell lysate, which is a suspension made by breaking open cells and removing membranes and other cellular debris. The remaining extract retains the crucial machinery for protein production, including:
- Ribosomes: These molecular machines translate the mRNA code into proteins.
- Transfer RNAs (tRNAs): These adapter molecules carry specific amino acids and interact with the mRNA codons to link amino acids together during protein synthesis.
- Amino acids: The building blocks of proteins.
- Enzymes: Various enzymes are necessary for translation initiation, elongation, and termination.
Types of Cell-Free Systems:
- There are two main types of cell-free systems:
- Cell extract-based: These systems utilize lysates from various sources like E. coli, wheat germ, or rabbit reticulocytes. Each source has advantages and limitations depending on the desired protein.
- Purified enzyme-based: These systems employ purified components like ribosomes, enzymes, and tRNAs, offering more control over the reaction conditions.
Benefits of Cell-Free Systems:
- Speed and Simplicity: Compared to culturing living cells, cell-free systems offer a faster and more streamlined approach to protein synthesis.
- Scalability: Protein production can be easily scaled up or down by adjusting the reaction volume and components.
- Purity: Proteins synthesized in cell-free systems are typically free of contaminants present in whole cells.
- Direct Manipulation: The system allows for easier manipulation of reaction conditions. Researchers can introduce non-standard amino acids or modify reaction parameters for specific purposes.
Applications of Cell-Free Systems:
- Rapid Protein Production: This technique is valuable for research purposes, such as studying protein function or screening for protein activity.
- Membrane Protein Production: Cell-free systems can be advantageous for producing membrane proteins, which are often difficult to express in living cells.
- In Vitro Diagnostics: Cell-free protein synthesis can be used to develop diagnostic tools by expressing specific antigens for antibody detection.
- Biomanufacturing: This approach holds promise for the future production of complex proteins for various applications.
Here's a breakdown of protein purification using a histidine tag (His-tag) in a cell-free system:
Cell-Free Protein Synthesis with His-Tagged Protein:
- 1. DNA Construct with His-Tag:
- You start with a DNA construct encoding the protein of interest. This DNA sequence is often engineered to include a sequence encoding a string of histidine residues (His-tag) at either the N-terminus (beginning) or C-terminus (end) of the protein.
- 2. Cell-Free Protein Synthesis:
- The cell-free system, containing a cell extract with the necessary machinery for translation, is used to synthesize the protein from the DNA template.
- During translation, the ribosomes translate the mRNA sequence, incorporating the His-tag along with the desired protein sequence.
Protein Purification using His-Tag:
- 1. Immobilized Metal Affinity Chromatography (IMAC):
- This is the most common technique for purifying His-tagged proteins. It utilizes a chromatography column containing beads coated with nickel (Ni) ions.
- Histidine residues in the His-tag have a high affinity for nickel ions.
- 2. Binding and Washing:
- The cell-free reaction mixture containing the His-tagged protein is passed through the chromatography column.
- The His-tagged protein specifically binds to the nickel ions on the beads, while other proteins in the mixture flow through the column and are collected as waste.
- Washing steps are then performed to remove any nonspecifically bound molecules.
- 3. Elution:
- To elute (release) the purified His-tagged protein, a solution containing a higher concentration of a competing molecule, often imidazole, is introduced.
- Imidazole competes with the His-tag for binding to the nickel ions, causing the His-tagged protein to detach from the beads and be collected as the purified protein fraction.
Advantages of His-Tag Purification in Cell-Free Systems:
- Specificity: The His-tag provides a specific and efficient way to isolate the protein of interest from the complex cell-free extract.
- Single-Step Purification: IMAC is a relatively simple and fast technique, offering single-step purification from the cell-free reaction mixture.
- Compatibility: Cell-free systems are generally compatible with His-tag purification methods.
Considerations:
- Cleaving the His-Tag (Optional): In some cases, you might want to remove the His-tag after purification. This can be achieved using specific enzymes that cleave the linker sequence between the His-tag and the protein.
- Optimization: Depending on the specific protein and cell-free system used, some optimization of the IMAC conditions (e.g., imidazole concentration for elution) might be necessary.
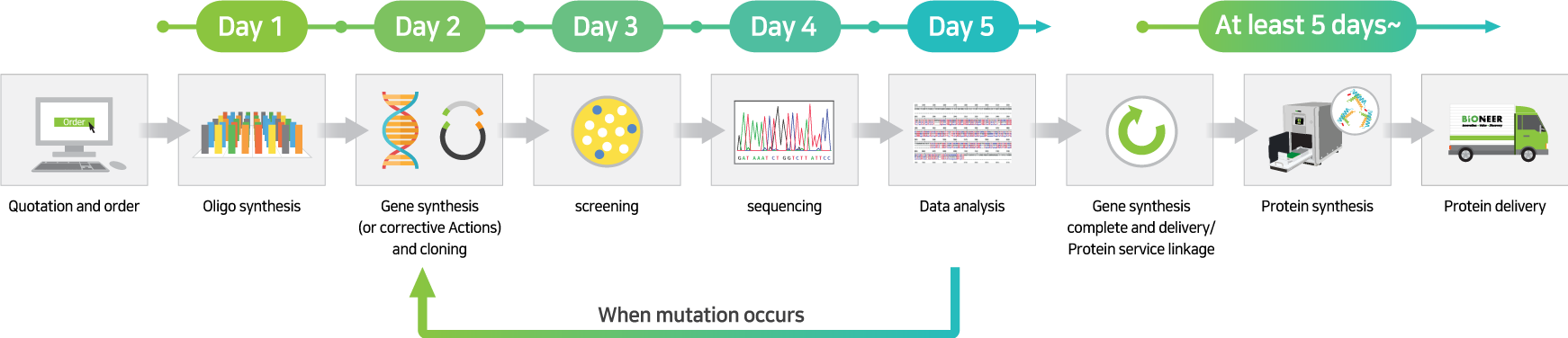
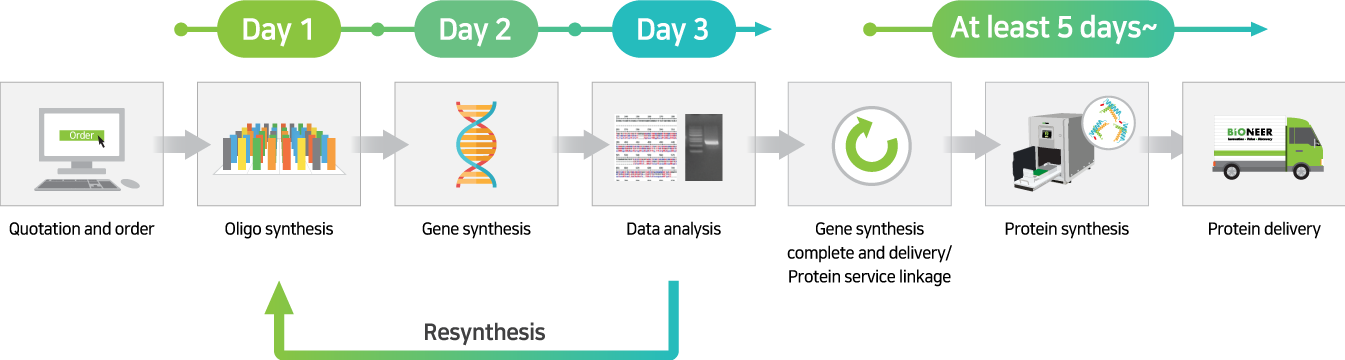
This is a service that synthesizes the gene for the protein you want to express, expresses the protein using ExiProgen™, purifies it, and then sends it to you.
By adding the gene coding for the protein you want to express to the protein synthesis kit, mounting it on the ExiProgen™ equipment, and operating it according to the manual, you can easily obtain purified proteins.
Standard Gene to Protein Service: This service proceeds with protein synthesis by adding the gene to the protein synthesis kit in the form of a plasmid cloned into an expression vector (pBT7-N-His or pBT7-C-His) after gene synthesis (300 bp to 3000 bp).
Cloning-free Gene to Protein Service: This service proceeds with protein synthesis by adding the gene in the form of a PCR product to a protein synthesis kit after AccuGeneBlock service (300 bp to 1000 bp).
Basically, the provided protein is supplied by dissolving it in the following buffer:
- Typical protein: 50mM Tris-Cl (pH7.6), 100mM NaCl, 1mM DTT, 0.1mM EDTA, 0.05% NaN3, 50% Glycerol
- Protein with disulfide bond: 50mM Tris-Cl (pH7.6), 100mM NaCl, 1mM DTT, 0.05% NaN3, 50% Glycerol
If you wish to use a special buffer, please write it down and send it when you submit your order.
Additional costs and time are required when proteins are supplied by dissolving them in a special buffer.