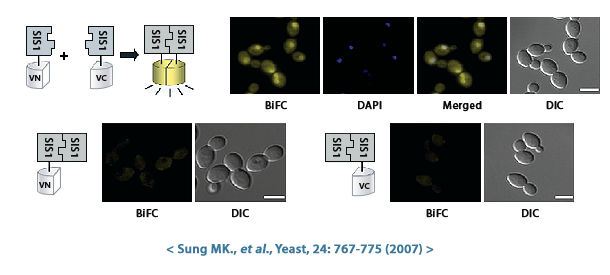
VENUS is a fluorescent protein derived from the yellow variant (YFP) of green fluorescent protein (GFP), commonly used in biological research. VENUS emits a brighter yellow fluorescent signal compared to GFP and is thus more easily detected in cells, and it also folds rapidly and matures quickly, allowing early visualization of protein localization. Additionally, VENUS has low phototoxicity, minimizing cellular damage caused by prolonged exposure to light during imaging experiments. In general, VENUS protein can be used to study protein localization, expression, and interaction in yeast cells.
- Intracellular location: By observing fluorescent VENUS signals in living cells, researchers can determine the cellular location of target proteins fused to VENUS. This helps us understand where proteins function within the cell.
- Expression: The intensity of the VENUS signal can be an indicator of the expression level of the target protein in the cell.
- Protein-protein interactions: By generating strains that co-express different VENUS-tagged proteins, researchers can utilize techniques such as bimolecular fluorescence complementation (BiFC) to identify proteins that interact with each other. When two proteins with interacting VENUS fragments come together, the VENUS fragments become close enough to form a complete fluorescent protein, producing a signal.
Bioneer has created haploid S.cerevisiae that expresses a fusion protein combining the VENUS fluorescent protein and the target protein, and is providing it as the S.cerevisiae Genome-wide VENUS-Fusion Library. VENUS protein is bound to the target protein in the form of an N-terminal fragment (VN) or a C-terminal fragment (VC). When the VN and VC fragments combine to form a complete VENUS protein structure, it displays fluorescence. And this VENUS-target protein fusion protein is expressed under the control of the target gene promoter.
The S.cerevisiae Genome-wide VENUS-Fusion Library is provided as VC- and VN-fusion libraries.
- S.cerevisiae VC-fusion library (5,552 strains): The C-terminal fragment of VENUS fluorescent protein (VC) is conjugated to the target protein, and was created in a haploid S.cerevisiae strain of MATa (mating type).
- S.cerevisiae VN-fusion library (5,809 strains): The N-terminal fragment of VENUS fluorescent protein (VN) is bound to the target protein, and was constructed in a haploid S.cerevisiae strain of MATα (mating type).
Diploid yeast and haploid yeast differ in the number of chromosomes they have.
- Diploid yeast: Diploid yeast cells have two sets of chromosomes (2n). In the context of yeast genetics, this usually means that there are two copies of each chromosome, one inherited from each parent.
- Haploid yeast: Haploid yeast cells have a single set of chromosomes (n). Haploid yeast cells usually arise through a mating event or specific genetic manipulation. Haploid yeast can be made into diploid yeast through sexual reproduction depending on the mating type.
Saccharomyces cerevisiae, also known as baker's yeast, reproduces sexually and has two mating types: MATa and MATα. This mating type is determined by a single mating type locus (MAT locus) located on chromosome 3.
- MATa type: This type contains MATa1 and MATa2 gene sequences in the MAT locus.
- MATα type: This type contains the MATα1 and MATα2 gene sequences in the MAT locus.
S.cerevisiae with different mating types reproduce sexually in the following manner:
- Yeast cell contact: Haploid cells of opposite mating types (MATa and MATα) must first come into physical contact through a process called agglutination.
- Signal transmission: Pheromones are secreted according to each mating type. MATa cells secrete a pheromone called a-factor, and MATα cells secrete α-factor. These pheromones bind to specific receptors of the opposite mating type and trigger a signaling cascade.
- Cell fusion: The pheromone signaling pathway induces cell wall remodeling and the formation of a fusion bridge between two cells.
- Nuclear fusion: The cell wall is ultimately completely destroyed and the two haploid nuclei fuse to form a diploid cell.
And the resulting diploid yeast cell receives one chromosome from each parent and has two sets of chromosomes. This haploid yeast mating technology can be used in genetic research, spore analysis, fermentation strain optimization research, etc.
Bimolecular fluorescence complementation (BiFC) is a technique used to study protein-protein interactions (PPI) in living cells. The BiFC assay principle and method using the S.cerevisiae Genome-wide VENUS-Fusion Library are as follows.
- VENUS fusion library is divided into S.cerevisiae VC-fusion library and S.cerevisiae VN-fusion library. In the VC-fusion library, the C-terminal fragment of VENUS fluorescent protein (VC) is bound to the target protein, and in the VN-fusion library, the N-terminal fragment of VENUS fluorescent protein (VN) is bound to the target protein. Both libraries were created using haploid S.cerevisiae, but the mating types are different: MATα (VC fusion library) and MATa (VN fusion library), respectively.
- Select the two types of target proteins A and B whose interaction you want to check, and then select strains expressing target proteins A and B bound to non-identical VENUS ends. For example, select a VC-protein A expression strain from the VC-fusion library and a VN-protein B expression strain from the VN-library.
- Sexual reproduction is possible because the VC- and VN-fusion library S.cerevisiae mating types are different into MATα and MATa, respectively. Therefore, we perform hybridization between VC- and VN-fusion library strains. In the diploid yeast cells created at this time, both target proteins A and B are present, combined with the VENUS fluorescent protein of the C-terminal and N-terminal fragments, respectively.
- If Protein A and Protein B naturally come close and the interaction is strong enough, the VN and VC fragments that were bound together will reassemble to form the complete fluorescent VENUS protein. This reassembled VENUS emits a yellow fluorescent signal, indicating a potential interaction between protein A and protein B. Conversely, even if the two proteins interact, if the interaction is weak or transient, the VN and VC fragments may not be close enough to display fluorescence.
S.cerevisiae grows at a temperature of 30℃, and the main medium composition is as follows:
1. YPD for general culture and maintenance medium
| Component |
Amt |
Final conc. |
| Yeast extract |
10 g/L |
1.0% w/v |
| Peptone |
20 g/L |
2.0% w/v |
| Glucose |
20 g/L |
2.0% w/v |
| Solid media is made by adding 2% Bacto Agar. |
2. SC medium for auxotrophic marker selection
- SC-Ura (S.cerevisiae VN-Fusion library selection): various additions to SD medium (Ade, Leu, Trp, His, Arg, Met, Tyr, Ile, Lys, Phe, Glu, Asp, Val, Thr, Ser) are added.
- SD-Leu (S.cerevisiae VC-Fusion library selection): various additions to SD medium (Ade, Ura, Trp, His, Arg, Met, Tyr, Ile, Lys, Phe, Glu, Asp, Val, Thr, Ser) are added.
- SD medium (Synthetic minimal glucose medium)
| Component |
Amt |
Final conc. |
| Bacto-yeast nitrogen base (w/o a.a) |
6.7 g/L |
0.67% w/v |
| Glucose |
20 g/L |
2.0% w/v |
| Solid media is made by adding 2% Bacto Agar. |
- Various additions for SC medium
| Constituent |
Final conc. (mg/L) |
Stock (g) per 100 ml |
Amt. of stock (ml) per L |
| Adenine sulfate |
20 |
0.2 |
10 |
| Uracil |
20 |
0.2 |
10 |
| L-Tryptophan |
20 |
1 |
2 |
| L-Histidine-HCl |
20 |
1 |
2 |
| L-Arginine-HCl |
20 |
1 |
2 |
| L-Methionine |
20 |
1 |
2 |
| L-Tyrosine |
30 |
0.2 |
15 |
| L-Leucine |
60 |
1 |
6 |
| L-Isoleucine |
30 |
1 |
3 |
| L-Lysine-HCl |
30 |
1 |
3 |
| L-Phenylalamine |
50 |
1 |
5 |
| L-Glutamic acid |
100 |
1 |
10 |
| L-Aspartic acid |
100 |
1 |
10 |
| L-Valine |
150 |
3 |
5 |
| L-Threonine |
200 |
4 |
5 |
| L-Serine |
400 |
8 |
5 |
The S.cerevisiae Genome-wide VENUS-Fusion Library is a collection of S.cerevisiae that express fusion proteins that combine the N-terminal fragment (VN) or C-terminal fragment (VC) of the VENUS protein with a target gene. The S.cerevisiae Genome-wide VENUS-Fusion Library consists of VC-fusion library and VN-fusion library depending on the VENUS fragment bound to the target protein. Both libraries used the S.cerevisiae TAP-tagged collection and were created by inserting a DNA cassette through homologous genetic recombination.
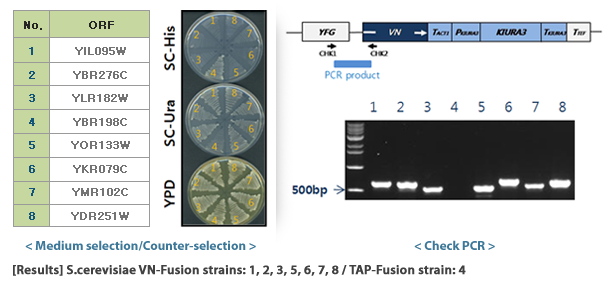
- Select target protein: Select the target protein to bind to VC or VN and check related strains in the S.cerevisiae TAP-tagged collection.
- DNA cassette preparation: Prepare a DNA cassette to create a yeast strain that expresses a fusion protein combining VC or VN with a target protein through homologous recombination. The F2CORE and R1CORE sequences used to create the TAP-tagged collection exist on both side regions of the DNA cassette, and between them, VC or VN sequences and auxotrophic marker genes for selective selection of yeast are inserted. As an auxotrophic marker, the KIURA3 (uracil) gene was used in the VC-fusion library and the LEU2 (leucine) gene was used in the VN-fusion library.
- Yeast transformation: DNA cassettes are introduced into yeast cells using transformation technology. The DNA cassette that enters the yeast cell recognizes the F2CORE and R1CORE sequences located near the target gene on the genome and is then inserted into the corresponding location. At this time, TAP-HISMX6, which was present in the TAP-tagged collection, disappears from the yeast genome and is replaced by the VENUS protein fragment of the DNA cassette and the auxotrophic marker.
- Selection of VENUS fusion strains: Transformed yeast is grown on auxotrophic medium and only yeast with the DNA cassette introduced into the genome are selected. Finally, all VENUS fusion strains are prepared as one set and provided as a library. In selected yeast, the target protein is expressed in a form linked to VC or VN, and expression is controlled by the target protein gene's own promoter.
Agar type and glycerol type are storage conditions for cultured yeast.
- Agar type: Agar type is a method of cultivating and storing yeast strains on a pre-prepared solid medium rich in nutrients. Agar serves to solidify the liquid medium. Agar type strains are cultured at 30℃ and stored at 4℃ to inhibit growth.
- Glycerol type: Glycerol type is a storage condition that allows liquid cultured strains to be cryopreserved (-70℃) by treating them with 30% (v/v) glycerol. Glycerol binds to water molecules within cells and acts as a cryoprotectant that protects cells by suppressing ice crystal formation even at low temperatures.
When ordering yeast strains from Bioneer, you can choose between agar type and glycerol type as the shipping type below.
- Agar type: Rich medium + supplements solid medium (2.0 microtube)
- Glycerol type: Rich medium + supplements + 30% glycerol liquid medium (2.0 microtube)
| Cat. No. |
Pack size |
Shipping temperature |
| Width (cm) |
Height (cm) |
Length (cm) |
Product weight (kg) |
Weight incl. dry ice (kg) |
Total weight (kg) |
| M-1010 (Agar type) |
22.5 |
16.5 |
5.8 |
0.158 |
- |
0.158 |
Room temperature |
| M-1010 (Glycerol type) |
5.4 |
4.5 |
5 |
0.049 |
10 |
10.049 |
Frozen |
A representative S.cerevisiae genome database can be found at the Saccharomyces Genome Database (SGD, https://www.yeastgenome.org).
SGD provides free, comprehensive information on the molecular biology and genetics of S.cerevisiae. Additionally, information can be obtained from:
There are two main methods for finding the human ortholog genes of S.cerevisiae genes:
1. Use SGD
SGD, the leading S.cerevisiae genome database, provides resources for finding human ortholog genes. Here's how to use it:
- Step 1: Go to the gene page for the S.cerevisiae gene of interest in SGD. You can search for genes by name or gene ID using the search box.
- Step 2: Click the “Homology” section at the top right of the results page.
- Step 3: If you enter “Homo sapiens” in the filter table under “Homologs” located at the middle of the page, you can check the human ortholog gene of the gene.
2. Sequence similarity-based search
If you cannot find a human ortholog gene for a S.cerevisiae gene, you can utilize a sequence similarity search tool such as BLAST (Basic Local Alignment Search Tool). A typical method of using NCBI BLAST is as follows:
- Step 1: The DNA or protein sequence of the S.cerevisiae gene of interest must be known. This information can be obtained through research or checked using gene ID in the SGD or NCBI databases (https://www.ncbi.nlm.nih.gov/search/).
- Step 2: After accessing the following NCBI BLAST page (https://blast.ncbi.nlm.nih.gov/Blast.cgi), type blastn, blastp, or blastx to use your DNA sequence (nucleotide) or protein sequence. Then select “homo sapiens” in the “Organism” tab and click BLAST at the bottom of the screen.
- Step 3: In BLAST results, the human ortholog sequence with the highest sequence similarity to the S.cerevisiae sequence is located at the top of the results with the lowest E-value.