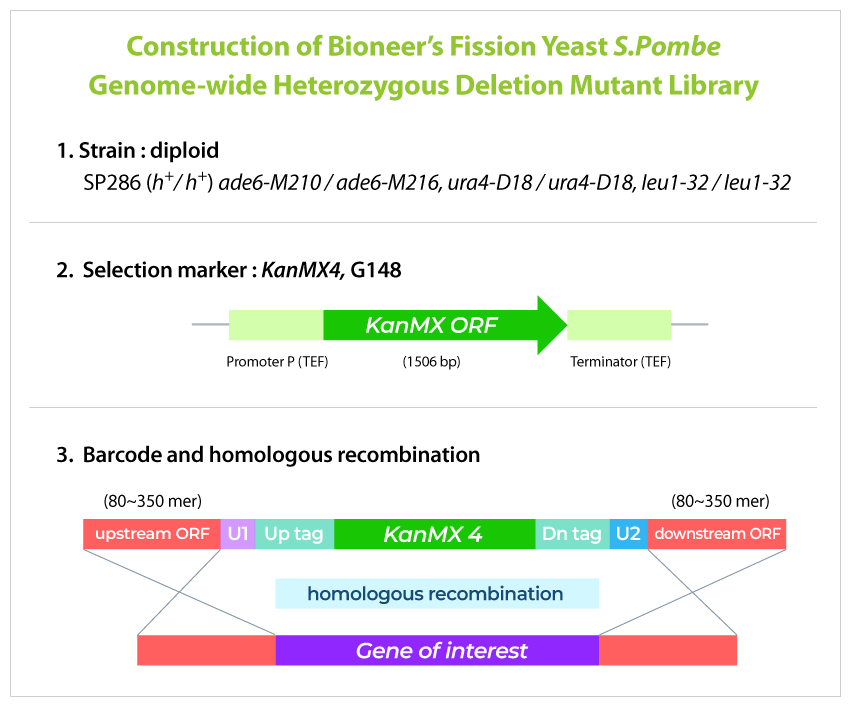
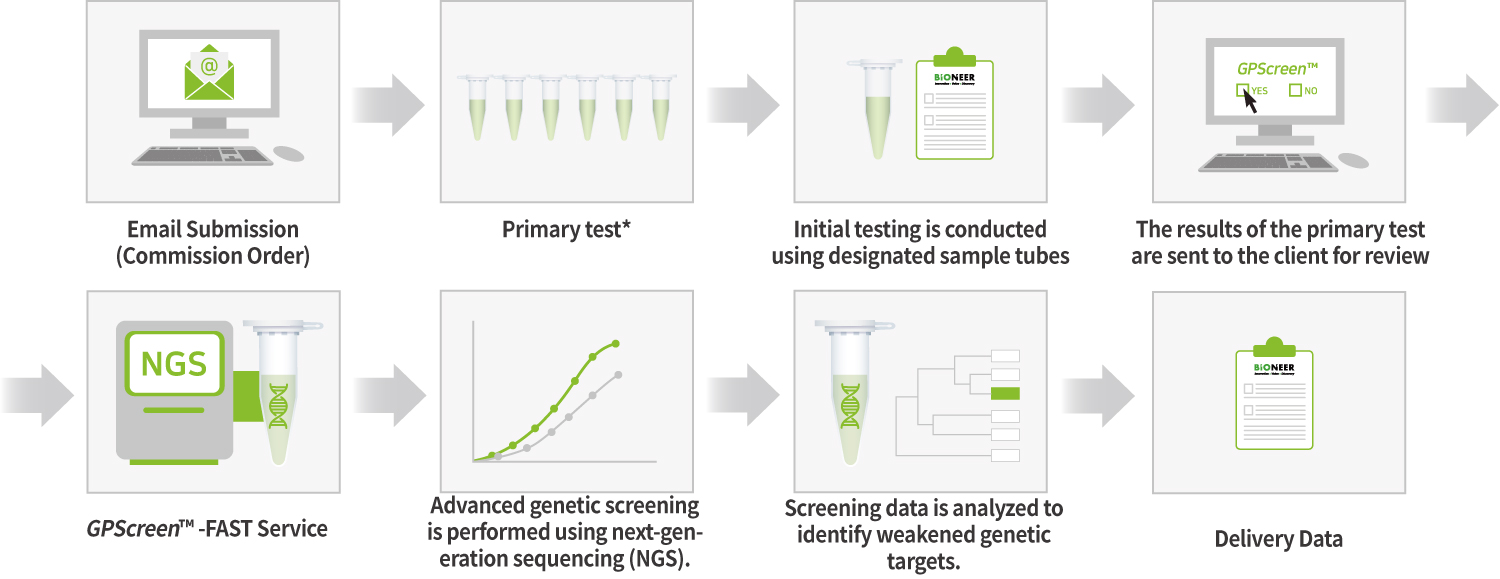
Yeast genome-wide knockout library is the world's only fission yeast gene deletion library developed through joint research between the Korea Research Institute of Bioscience and Biotechnology (KRIBB) and the Royal Cancer Institute (Cancer Research UK) (Nature Biotech., 2010). The library is composed of mutant fission yeast (S. pombe) lacking the function of target yeast genes, and targets approximately 98.5% of the genes of the entire fission yeast genome. Gene function-deficient variants were created by inserting an antibiotic resistance gene and a barcode unique to the variant instead of the target gene using the homologous gene recombination method.
GPScreenTM-FAST balances the growth of all variants that make up the library and then pools them for service.



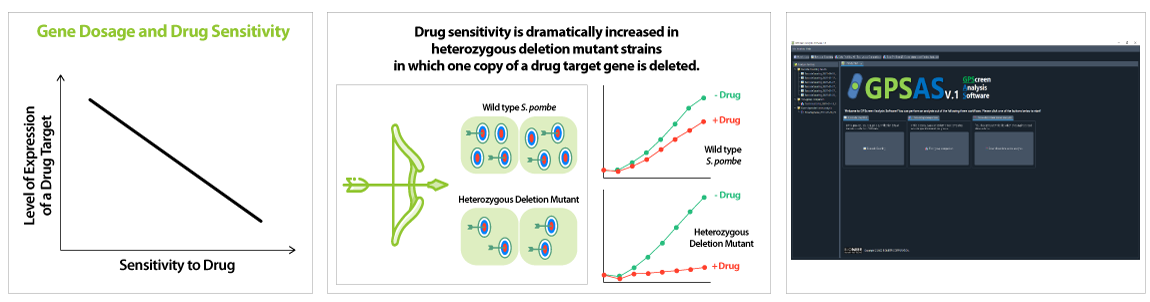
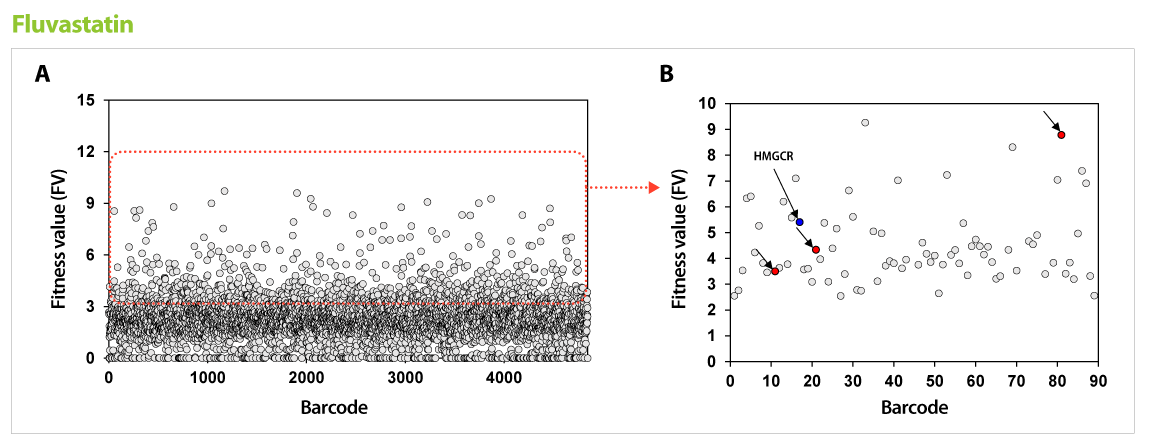
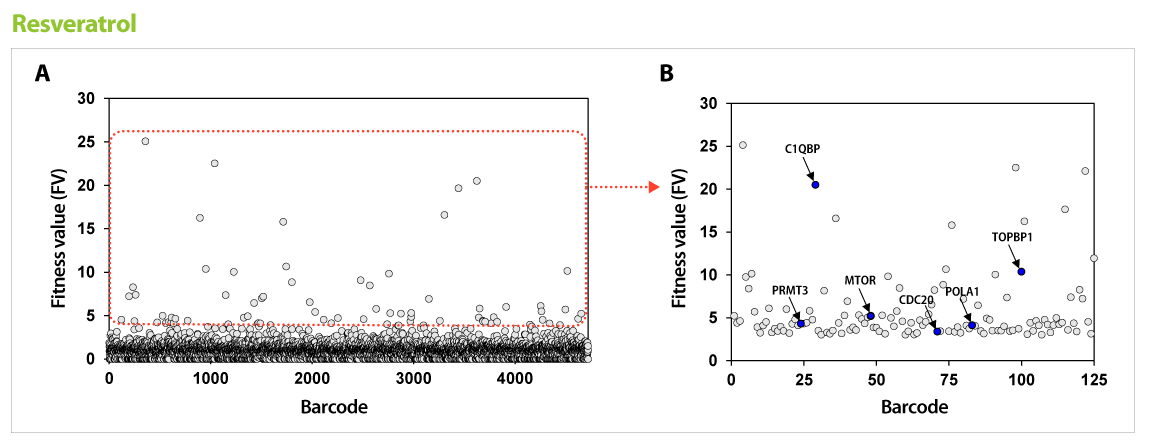
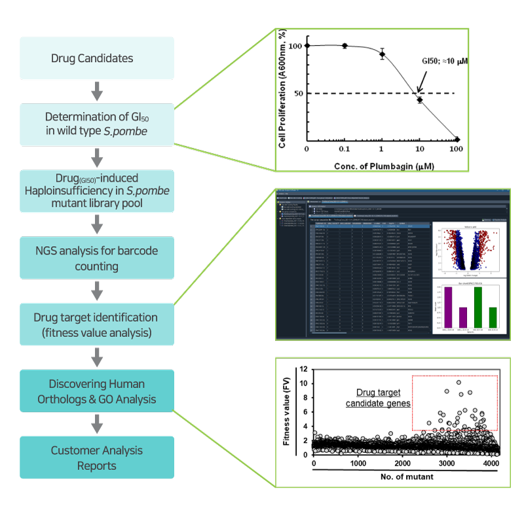
▶ Using the drug action point discovered via GPScreen™ , target validation studies are possible on human cells.
IMethods for Validating Drug Targets in Human Cells
Bioneer has about 20,000 siRNA libraries for human genes.
By using the human orthologs of the identified target candidates from GPScreen™, gene knockdown can be analyzed to verify the gene in human cells using the siRNA library.
1) Analysis of human homologous gene expression regulation by drugs in human cells
Figure 1. Photos and the graphs showing the inhibition of POLG expression by Sunitinib in the Hela human cell lines
(A) To measure the cellular responses of Hela cells to sunitinib, the cells were spread onto m-slide 8-well plates and treated with 0 µM or 5 µM of sunitinib for 48 h. Then, the level of POLG proteins was observed with immunocytochemistry using an anti-POLG antibody and goat anti-rabbit IgG-TRITC antibody in fluorescent confocal microscope.
(B) Hela cells were treated with 0 µM or 5 µM of sunitinib for 48 h, and POLG mRNA levels in the treated cells were measured by qRT-PCR.
2) Analysis of inhibitory effect of human homologous gene expression using BIoneer's human siRNA library
Figure 2. Photos and the graphs showing the knockdown of POLG dramatically increased the cytotoxicity by sunitinib in a human cell line (Hela cells).
(Upper panel) After the treatment of siRNAs (5 nM, 48 h), the cells were further treated with sunitinib (5 µM) for 48 h, and the cell mass was measured by SRB assay.
(Bottom panel) The cells that have went through SRB assay were observed under a microscope.
IIApplication of GPScreen™ technology to derive new drug development candidates
In addition, GPScreen™ technology is also available for the screening of safer compounds between drug candidates in the early stages of new drug development for comparison. When this technology was used for analysis of target profiles at the genome level, toxic drugs showed a broader target spectrum than non-toxic drugs. It represents the toxicity profile of the drug in the human body and can be used effectively for drug prioritization for the safest drug among the same kind of candidate substances.
Figure 3. Targeted spectrum comparison after GPScreen™ each toxic and nontoxic drugs